John Terhorst, Ph.D.
Chemistry Instructor/Lecturer
Ph.D., Chemistry, Yale University, 2011
M.S., Chemistry, Yale University, 2008
B.S., Chemistry (Honors) & Biology, University of Redlands, 2006
Joined Oregon State University Faculty, September 2023
Joined California Institute of Applied Technology Faculty, April 2023
Joined University of Redlands Faculty, August 2020
Wyzant Chemistry Tutoring, 2012-2023
Vanguard University Faculty, 2012-2023
Ecampus Excellence in Online Teaching and Student Engagement Award, Oregon State University, 2025
Dox Research Fellowship, Yale University, 2009
Distinguished Chemistry Fellowship, Yale University, 2006-2011
Robert D. Engel Award, University of Redlands, 2006
Edmund C. Jaeger Award, University of Redlands, 2005
Tutoring
Let me help you master chemistry!
I am a college instructor and private tutor with 18 years of teaching experience. In these roles I teach college classes and engage with students to help nurture a passion for and excitement about science. I offer one-on-one and group-based chemistry lessons online from my home in Redlands, California. My hours are flexible and I can accommodate most requests. The subjects I most often teach are introductory, honors, A.P., general, and organic chemistry.
I am patient with students and do my best to promote a relaxed, productive, and respectful working relationship. I am willing to take on students who are struggling or those who are looking to get ahead in their studies. I can help you with:
- Introductory, Honors, and AP Chemistry
- General Chemistry I and II
- Organic Chemistry I and II
- ACS Exam Prep
- SAT II Chemistry Subject Exam Prep
- Spectroscopy (IR, MS, and NMR)
- Computational Chemistry
Booking
My rate is $125/hour for all lessons. Contact me or schedule a lesson.
Featured Testimonials
"John has been a game changer for my daughter! He has helped her to feel so much more at ease and enjoy her chemistry class by working with her to strengthen her understanding of each unit. Since starting to work with John, her test grades have shot up, and she is confident in her ability to do well in chemistry. John is incredibly knowledgeable of the material, and patient and explains difficult concepts easily as well. We will definitely continue to work with John, and highly recommend him to students at any level of chemistry."
Rosie, 51 lessons with John
"Awesome teacher! When my daughter failed her chemistry midterm I was looking for a qualified chemistry tutor. After going through several of them, I was quite disappointed because they focused on solving problems without sufficient explanation of the chemistry concepts. That's when I met John. He really took his time explaining the chemistry concepts, allowing my daughter to get a greater understanding of the concepts, and be confident knowing that she could solve difficult chemistry problems. My daughter jumped from failing her midterm to acing her final. Thanks again, John."
Don, 32 lessons with John
"John is an incredible tutor - his patient and calm demeanor makes for a comfortable learning environment and his easy to follow explanations provide a perfect foundation for further knowledge. He sets you up for success by giving you a way to approach problems and understand what each type is asking for. He works out each problem on a whiteboard and talks through each step-it is insanely helpful! It really allows for you to take things slow and understand what is going where. I used John as a tutor for organic chemistry, I'm a student at UC Irvine and I was in my third quarter of ochem. As a professor himself, John was able to answer ANYTHING I asked and related the ochem I had learned in my previous quarters back to what he was explaining. It really helped me to see the connections between information. Tutoring from John is that one-on-one extra office hour you so need with the professor but never get because of all the students. If you need help in chemistry - this is your guy, he is really really amazing."
Sophia, 5 lessons with John
"Fantastic Tutor - John helped me with stereochemistry and NMR, and he was the best organic tutor I have had. Not only did he help me complete the assignment, he also explained very important concepts and why things happen. He was extremely patient and helpful, and I would definitely recommend him as an organic tutor."
Gina, 17 lessons with John
"Early in my daughter's first term of Organic Chemistry it was apparent she was struggling. John is an amazing tutor and has been an excellent fit for her. He is very patient. He not only explains underlying principles but anticipates and prepares her for the next question she is likely to see on an exam. He is flexible and very generous with his time. With John's help my daughter completed the term with an A- and has started working with John again this term!"
Barbi, 12 lessons with John
"John tutored my daughter in AP Chem from November 2016 through May 2017 on a weekly basis. The AP Chem class was hands down the toughest class my daughter took in school. The year before she took Honors Chem and got As in the class without help. AP Chem was altogether a different story. Right away I hired her a highly recommended chemistry/physics tutor used by several of my friends. But after a month my daughter said he wasn't able to explain the concepts to her and had to look up on google many of the questions she had for him. When I looked for a replacement tutor, what drew me to John was his extensive credentials in chemistry. I am so glad I chose him. In the six months he tutored my daughter, she said he was always patient, that he always knew the answers to the questions she asked, and instead of just having her memorize things, he would walk through the steps to each problem and explain the ideas behind the concepts so she would really understand it. From my perspective, John has always been beyond professional in providing prompt responses and very fair and accurate time for his lessons. If you are in need of an excellent chemistry tutor who really knows the subject, give him a try, you will not regret it."
Julia, 24 lessons with John
"John is an amazing tutor. Without him, I would not have been able to continue taking Organic Chemistry. I was very close to dropping the class, but with John's help, I was able to continue and excel in the subject. I could ask him any type of organic chemistry (and even biochem) question, and he not only knows the answer, but explains the concept and details behind the problem. He never made me feel unintelligent for not knowing a specific answer, and would patiently work through each problem. Thanks to John, I have a better and more clear understanding of a highly complex subject, that I would not have been able to attain if I had done it on my own."
Stefani, 11 lessons with John
"The best decision I made during my time as an organic chemistry student was to start lessons with Dr. Terhorst. It was not a subject that came easily to me at all, but Dr. Terhorst was able to take any question or example I had from class material and break it down in a way that made sense. He emphasized the underlying reasoning behind concepts which made them much easier to remember. Dr. Terhorst also always had tables of information he had prepared (and explained in depth during lessons) that were much more organized than material that I received from from my class, making studying much more efficient. It is clear that he not only has a passion for the subject, but also for teaching. This goes a long way and I would urge anyone in an organic chemistry class to reach out to Dr. Terhorst."
Andrew, 23 lessons with John
Research
My research projects use computers to explore questions in modern chemistry that experimental methods often cannot answer. Specifically, we apply chemical theory, statistics, and informatics to study the relationship between the structure of organic molecules and their activity in biological systems. This "quantitative structure-activity relationship," or QSAR, allows us to design molecules with certain desired pharmacological properties, with the ultimate goal of developing therapeutic agents targeting infectious, inflammatory, and hyperproliferative diseases.
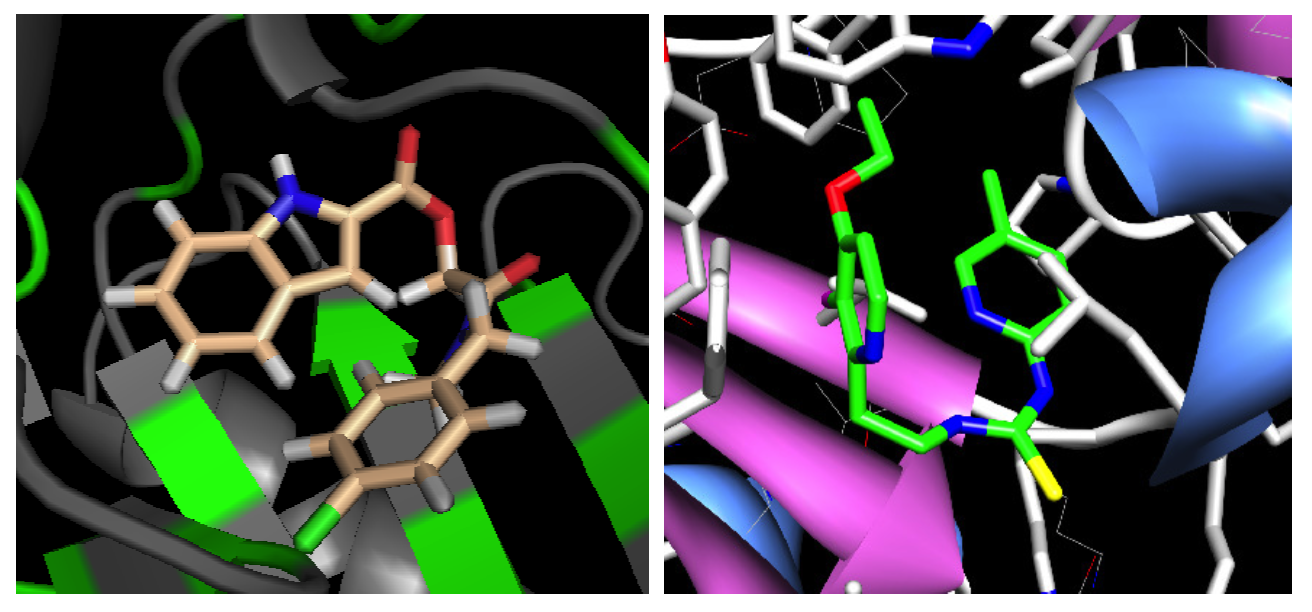
A Fragment-Based Search for HIV Inhibitors using Linear-Response Property Prediction
The foundation of any QSAR analysis begins with the fragment library: a database of molecular fragments and their associated geometric, physical, and chemical properties. This project involves design and population of a fragment library for discovery of inhibitors of HIV-1 reverse transcriptase, a key enzyme in the retroviral reproductive pathway. Research articles published in 2011 and 2012 indicate that heterocyclic cores connected by various linker groups can bind irreversibly to an allosteric site on HIV-RT, but novel structures such as these have not been widely studied in a systematic fashion. Our goal is to design a large set of heterocycles, linker groups, and derivatives, then to characterize them as individual fragments and as combinations thereof. Once the library is complete, we will use it to design drug candidates that should bind to our target based on complementary geometries, torsions, polarities, and hydrogen bonding; docking our candidates into the allosteric binding pocket of HIV-RT will then allow us to rank our results quantitatively.
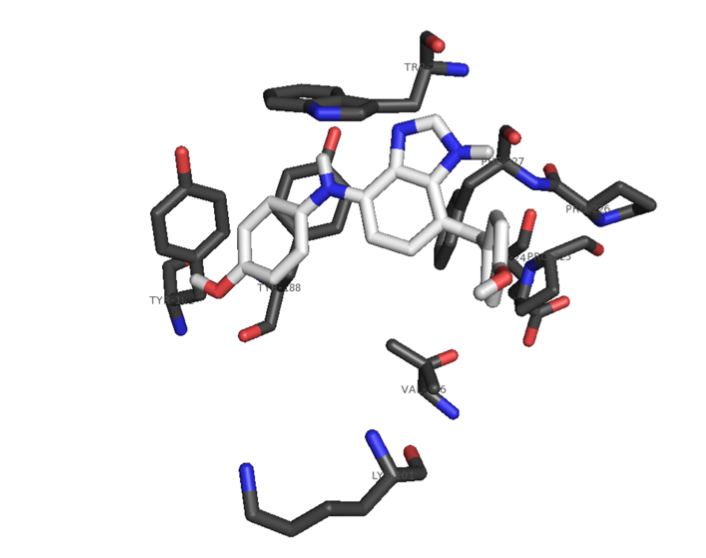
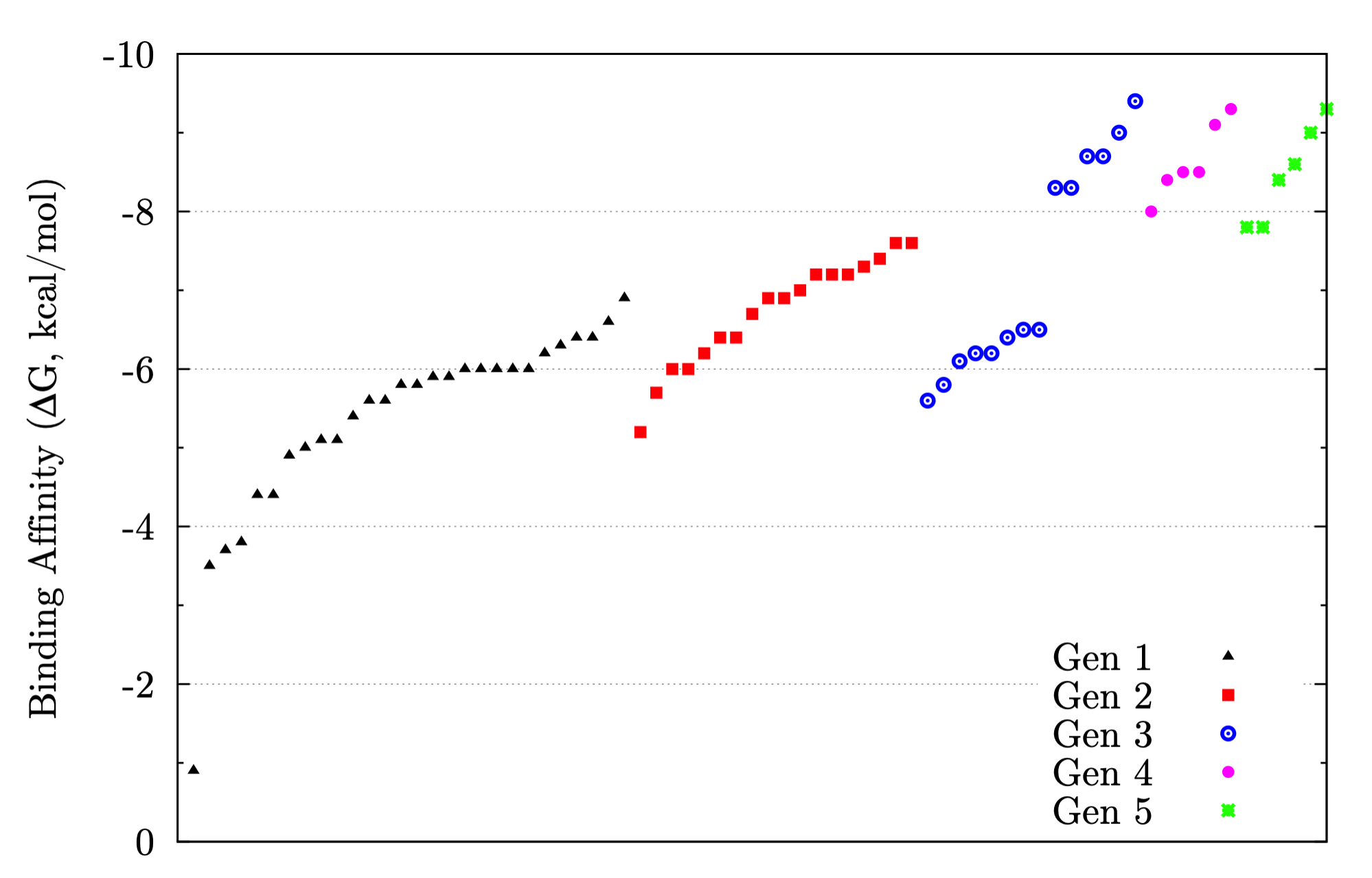
Continuum Solvation Models and Force Field Development for Computer-Aided Drug Design
My graduate work involved development of a novel method for rapid free-energy calculations with continuum solvent models.
In my dissertation, an implementation of the generalized Born / surface area (GB/SA) solvation model with free-energy perturbation (FEP), including an approximation used in calculating the total Born energy of the system, is presented. Our approximation is based on the assumption that a significant number of pairwise energy calculations may be omitted with little-to-no impact on the total change in energy of the system after a Monte Carlo move because the impact of a moving atom on the Born radius of a distant atom is small. Thus, we structured our implementation of GB/SA in such a way that the Born energy between an unmoving pair of atoms is only recalculated after a move if the Born radius of either atom has changed by more than a specified threshold since the last accepted move. Prior benchmarks demonstrated that existing GB/SA methodologies were insufficient for the purposes of calculating free energies of binding, and FEP simulations with GB/SA solvation were too computationally expensive to be used with any practicality. With our approximation, improved efficiency was achieved while affording minimal error: the influence of our approximation on accuracy of free energies of binding was negligible, with any error introduced by the approximation falling well below the statistical error of the Metropolis Monte Carlo algorithm, and speed-up of up to 62% was observed. The conclusion is that with our approximation, GB/SA is a viable solvent choice for FEP of large systems. Comparison between GB/SA and TIP4P in a substituent scan was quantitative to qualitative, with free energies of binding usually in agreement within 1 kcal/mol, producing the same substitution pattern on a drug candidate found to give high anti-retroviral activity as predicted by previous simulations with TIP4P explicit water.
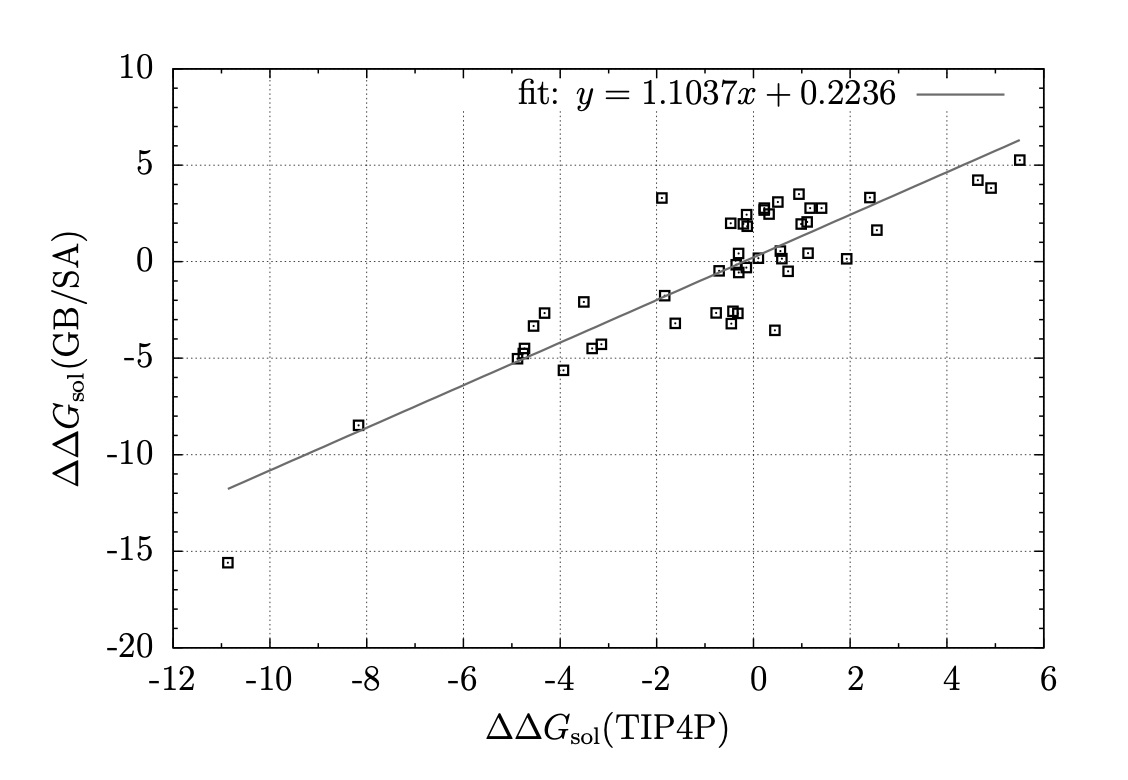
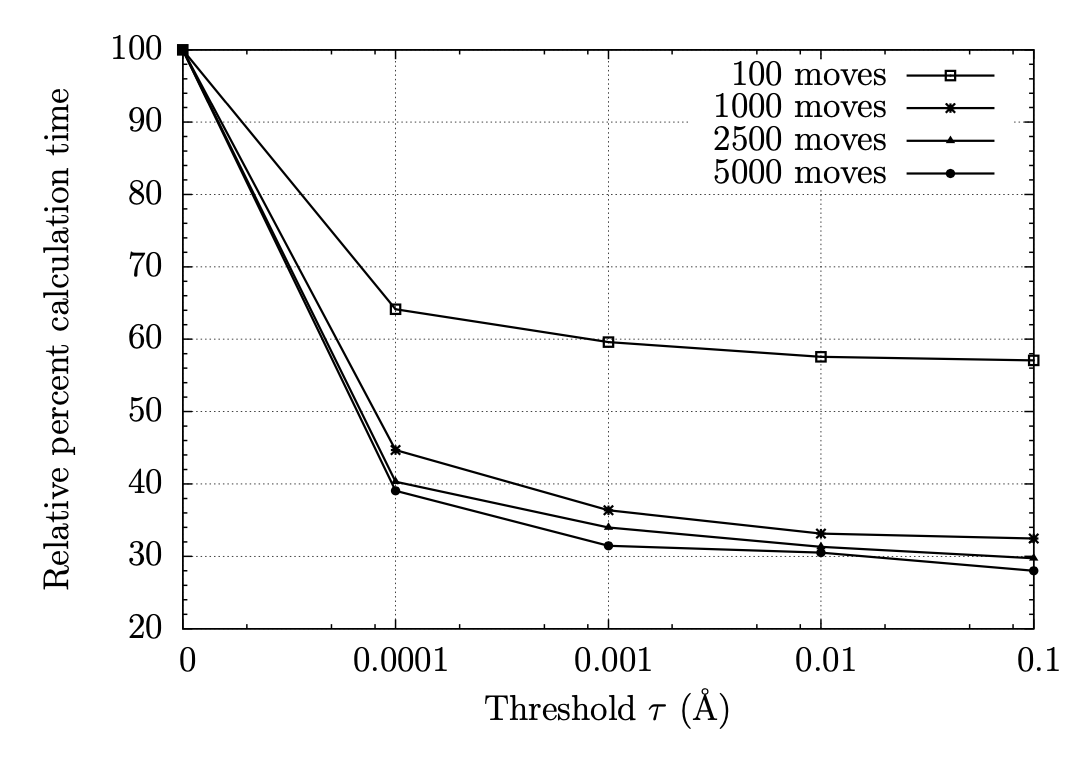
E/Z Energetics for Molecular Modeling and Design
Thermochemical data obtained from G3B3 quantum mechanical calculations are presented for 18 prototypical organic molecules that exhibit E/Z conformational equilibria. The results are fundamentally important for molecular design including the evaluation of structures from protein--ligand docking. For the 18 E/Z pairs, relative energies, enthalpies, free energies, and dipole moments are reported; the E-Z free-energy differences at 298 K range from +8.2 kcal/mol for 1,3-dimethyl carbamate to -6.4 kcal/mol for acetone oxime. A combination of steric and electronic effects can rationalize the variations. Free energies of hydration were also estimated using the GB/SA continuum solvent model. These results indicate that differential hydration is unlikely to qualitatively change the preferred direction of the E/Z equilibria.
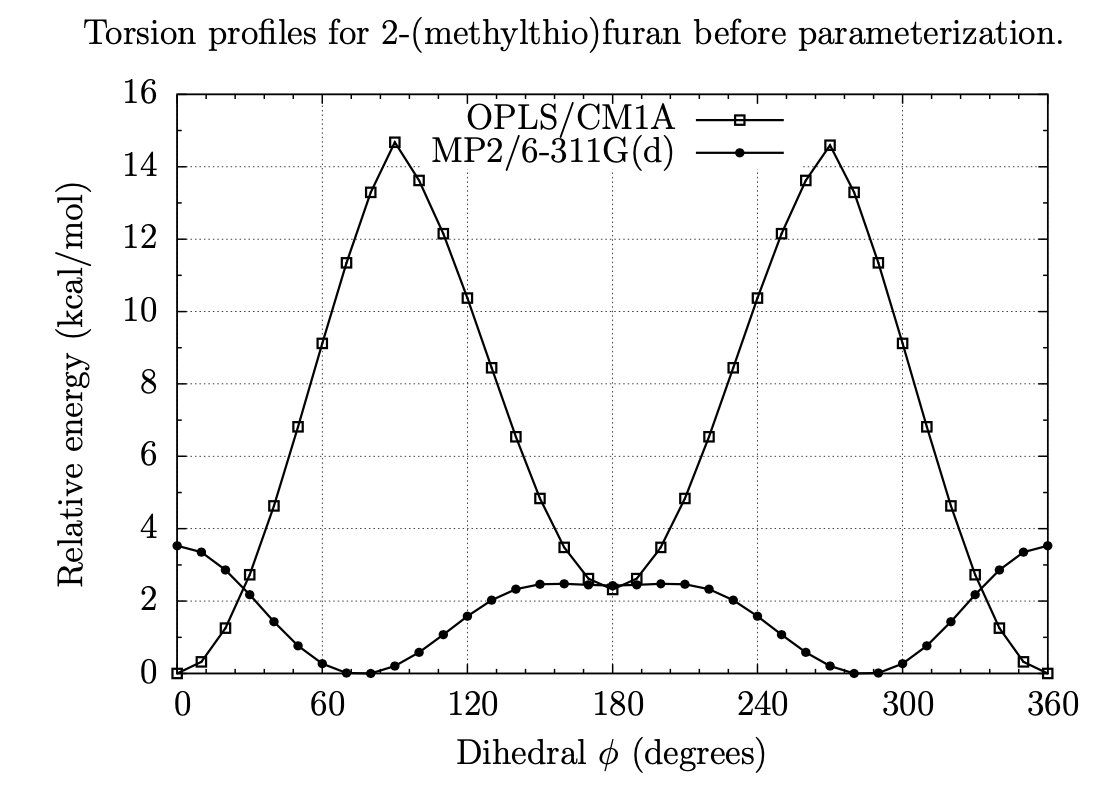
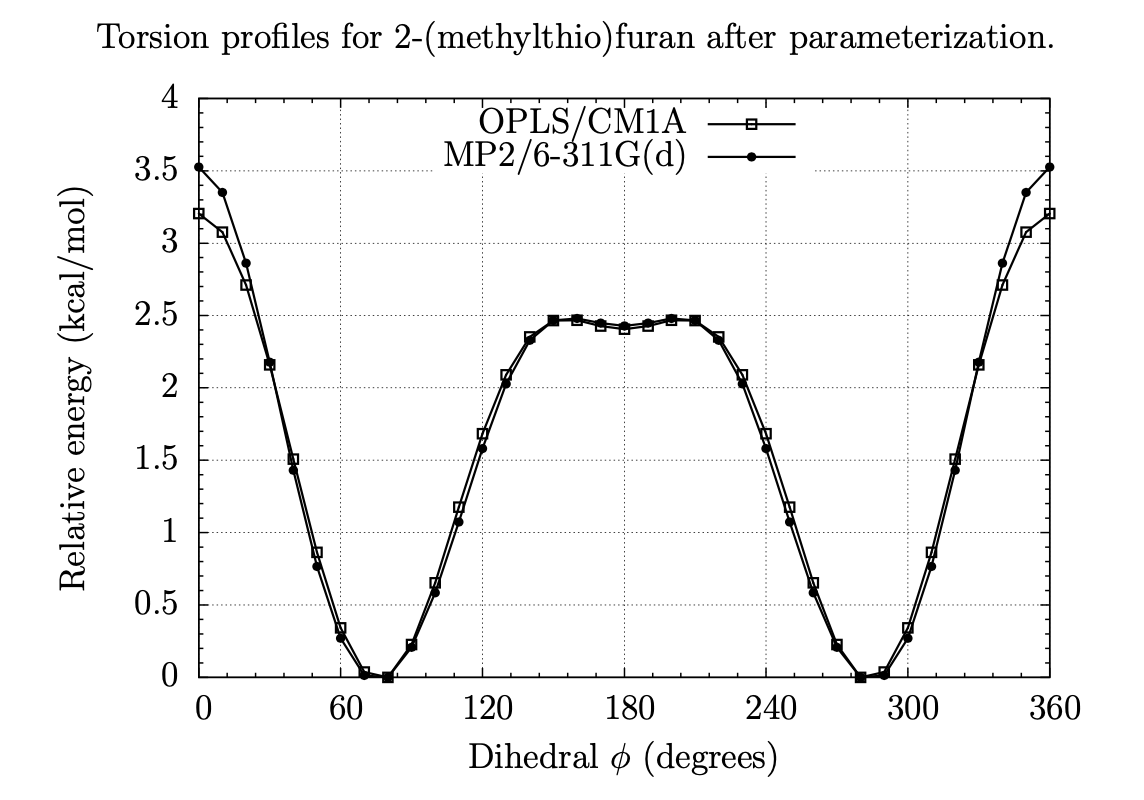
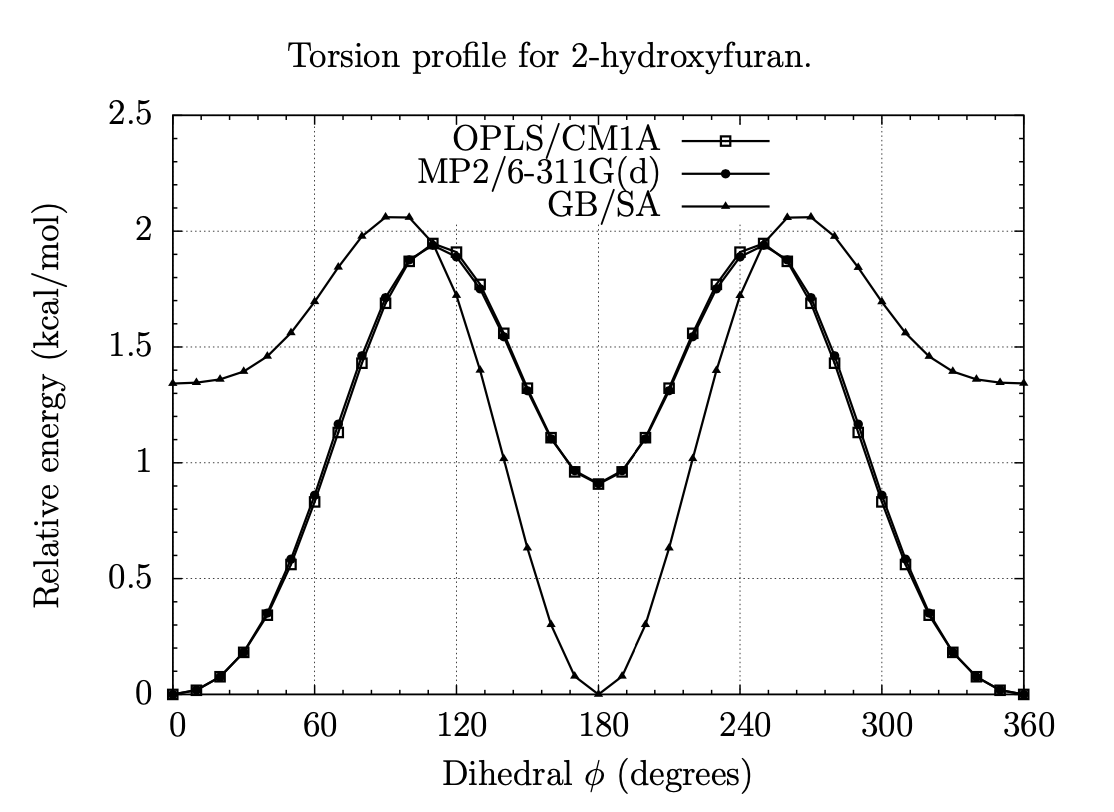
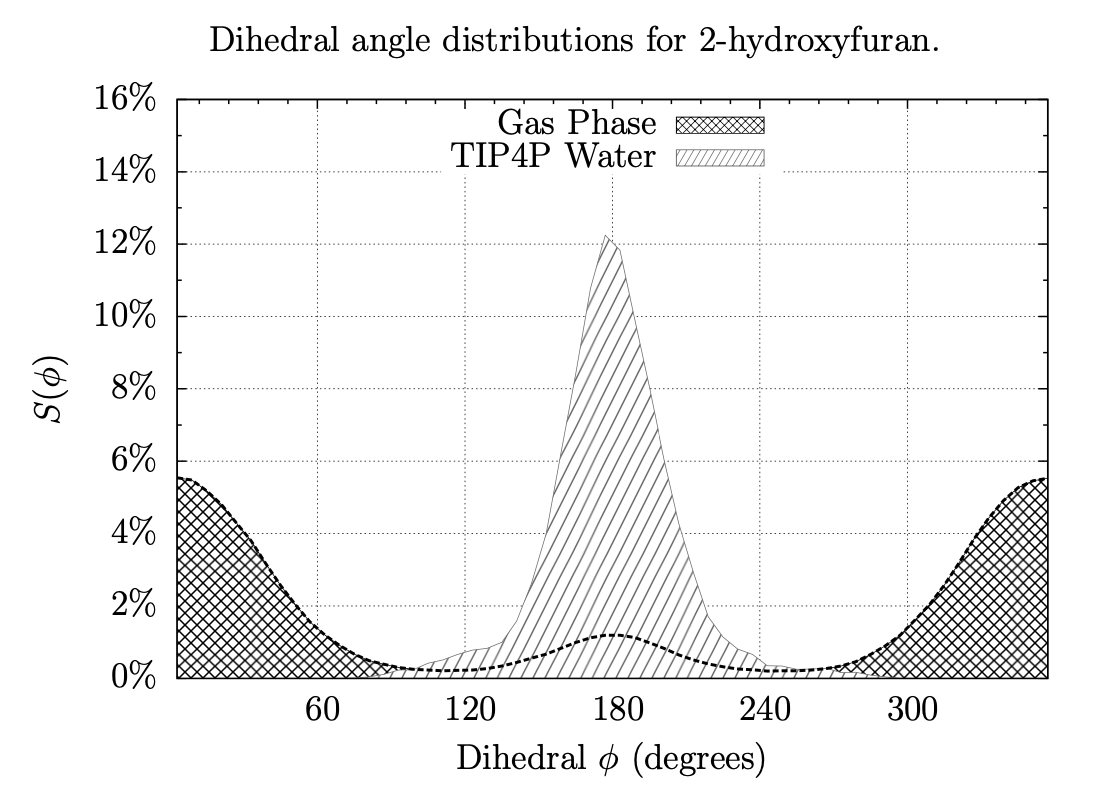
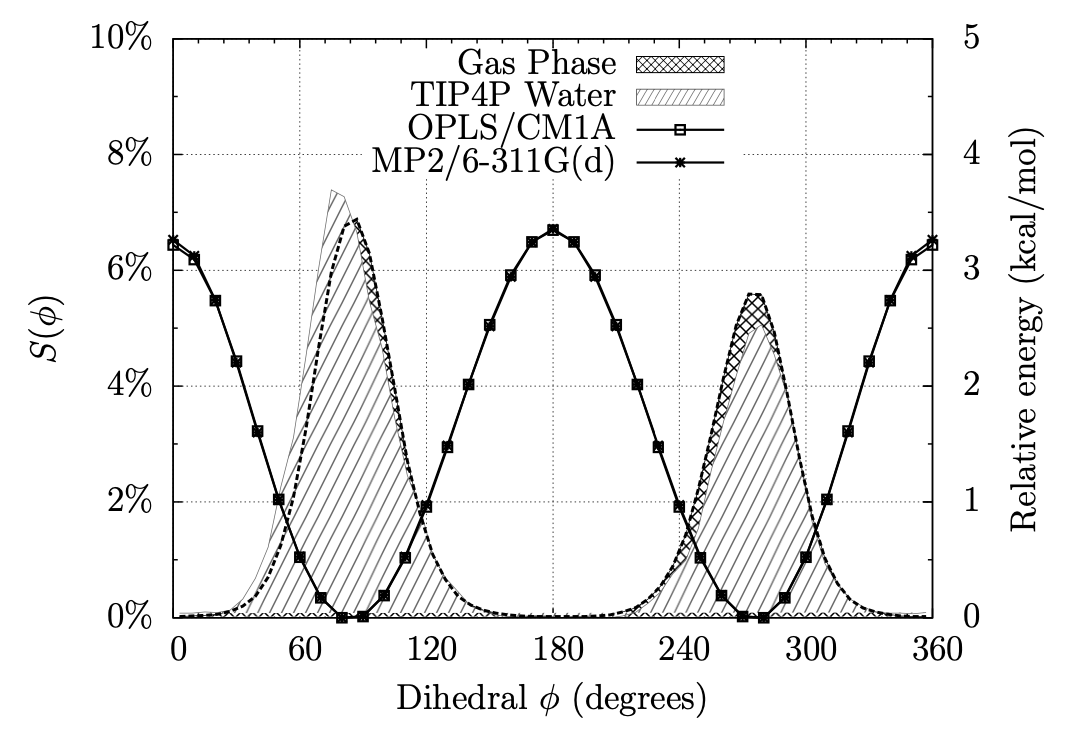
Exploring Dihedral Torsion Profiles with Implicit Solvent Models
Molecular mechanics are central to applications in computer-aided drug design. Of particular interest is the ensemble of conformations defined by one or more dihedral torsions within a given molecule. Often it is the case that these torsions give rise to unique molecular shapes, which can affect a potential drug's ability to bind (or not bind) to its intended target. For instance, if the conformation that is required for binding is too far uphill in energy from the molecule's native conformation, then binding is unlikely to occur; but if structural modifications can be made intelligently that can "lock" a molecule into its binding pose or otherwise coerce a molecule into a new low-energy conformation, binding can be enhanced. Thus, this project involves examining the dihedral torsion profiles of drug-like organic molecules in an aqueous environment to evaluate, among other things, the effects of solvation and substitution on molecular shapes. Our goal is to use chemical simulations to characterize the torsional energies of derivatives of benzene, pyridine, pyrimidine, pyran, furan, thiophene, and other heterocycles, with and without the effects of solvation by water; comparing the two sets can give insight into how the different classes of molecules conform under biological conditions and what energies are involved in their conformational equilibria.
Scientific Articles
1.
E/Z Energetics for Molecular Modeling and Design.
J. Terhorst and W. L. Jorgensen
J. Chem. Theory Comput. 2010, 6:9, 2762-2769.
doi:10.1021/ct1004017


2.
Simulations of Photopumping in Doubly Illuminated Liquid Membranes Containing Photoactive Carriers.
T. L. Longin, J. Terhorst, and C. Lang
J. Phys. Chem. B 2010, 114:48 15846-15858.
doi:10.1021/jp106802q



Complete Vita
Links

Last updated: 2023-03-29 05:33 GMT















